Efficient search, mapping, and optimization of multi‐protein genetic systems in diverse bacteria | Molecular Systems Biology

1 The ribosome binding site and 5 0 protein coding sequence control an... | Download Scientific Diagram

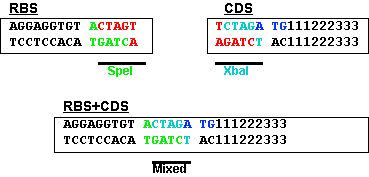
Efficient engineering of chromosomal ribosome binding site libraries in mismatch repair proficient Escherichia coli | Scientific Reports

Sequence logos of putative promoters, ribosome-binding sites (RBS) and... | Download Scientific Diagram
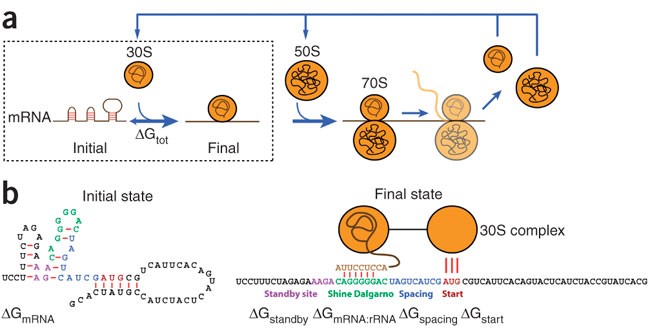
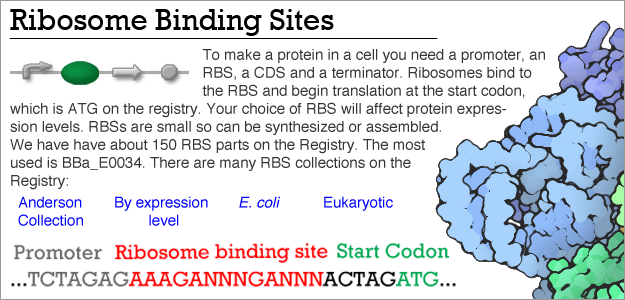
Automated design of synthetic ribosome binding sites to control protein expression | Nature Biotechnology
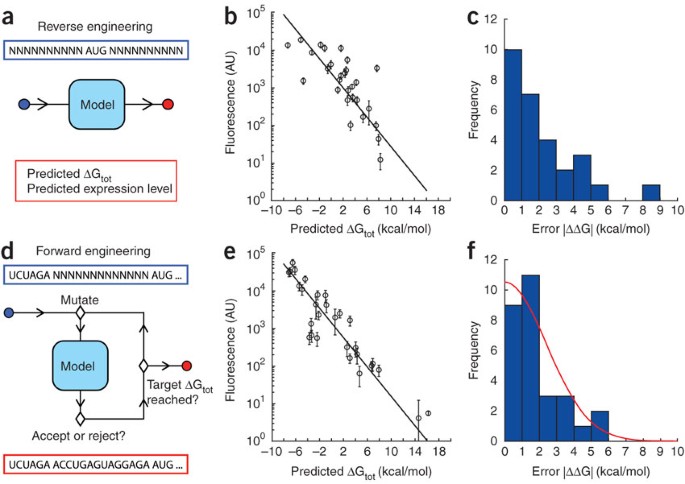
Automated design of synthetic ribosome binding sites to control protein expression | Nature Biotechnology
GitHub - hsalis/Ribosome-Binding-Site-Calculator-v1.0: The Ribosome Binding Site (RBS) Calculator can predict the translation initiation rate of a protein coding sequence in bacteria and design synthetic RBS sequences to rationally control the translation
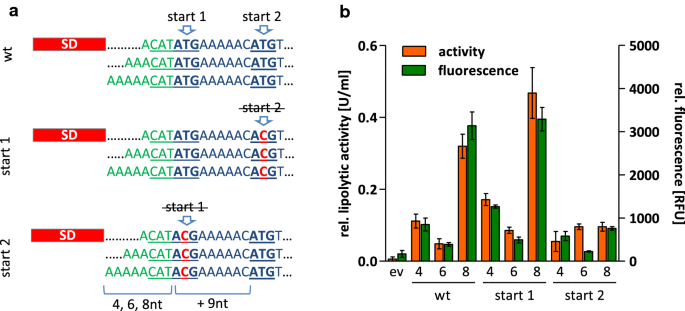
The length of ribosomal binding site spacer sequence controls the production yield for intracellular and secreted proteins by Bacillus subtilis | Microbial Cell Factories | Full Text
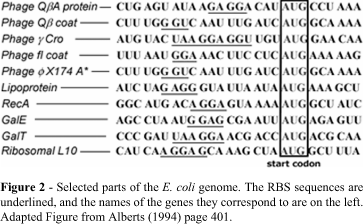
SciELO - Brasil - Ribosome binding site recognition using neural networks Ribosome binding site recognition using neural networks

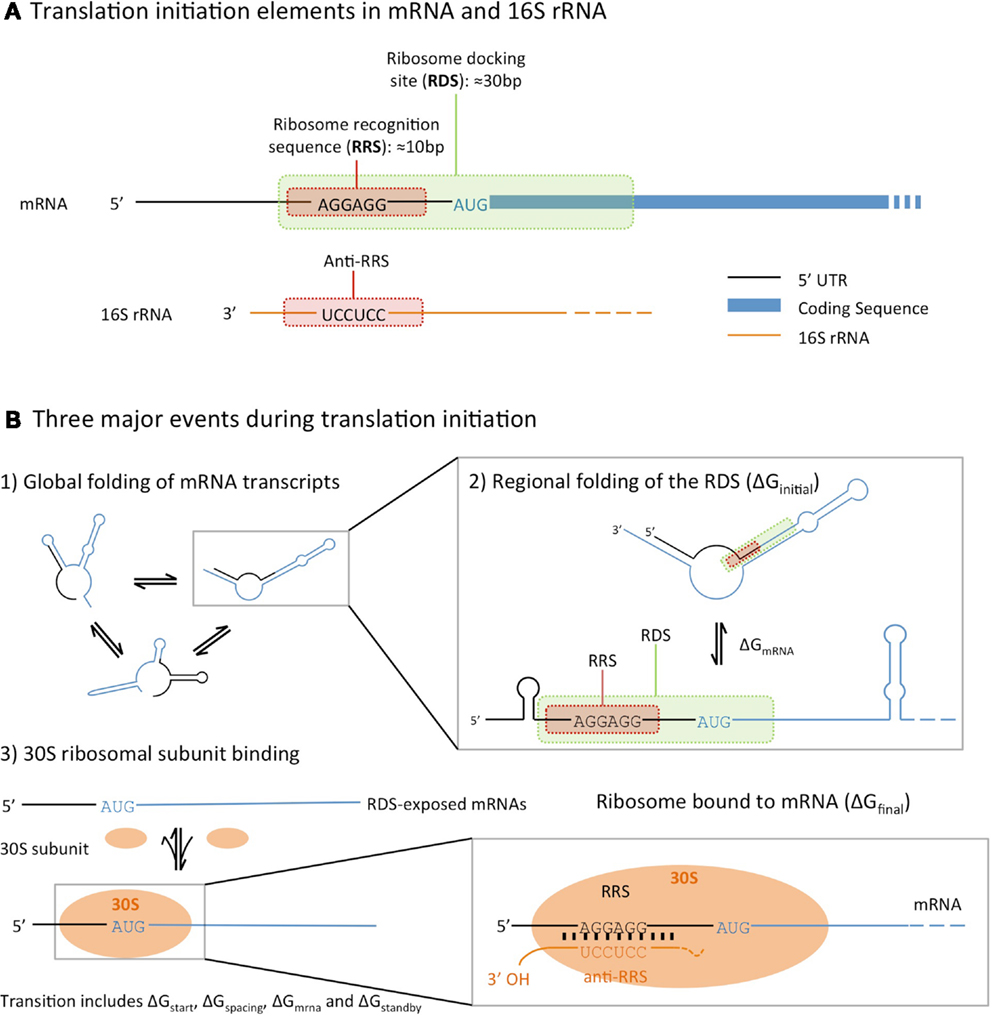
Deciphering the Rules of Ribosome Binding Site Differentiation in Context Dependence | ACS Synthetic Biology

What could be the possible distance of Ribosome Binding Site (RBS) from Translation Start Site? | ResearchGate

Deciphering the Rules of Ribosome Binding Site Differentiation in Context Dependence | ACS Synthetic Biology

Influence of the spacer region between the Shine–Dalgarno box and the start codon for fine‐tuning of the translation efficiency in Escherichia coli - Komarova - 2020 - Microbial Biotechnology - Wiley Online Library

i) DNA templates containing a T7 promoter (1), ribosomal binding site... | Download Scientific Diagram
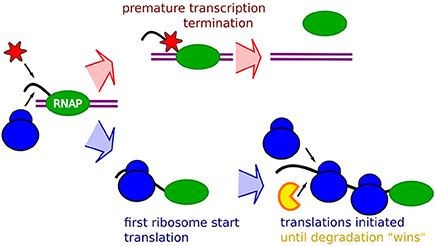
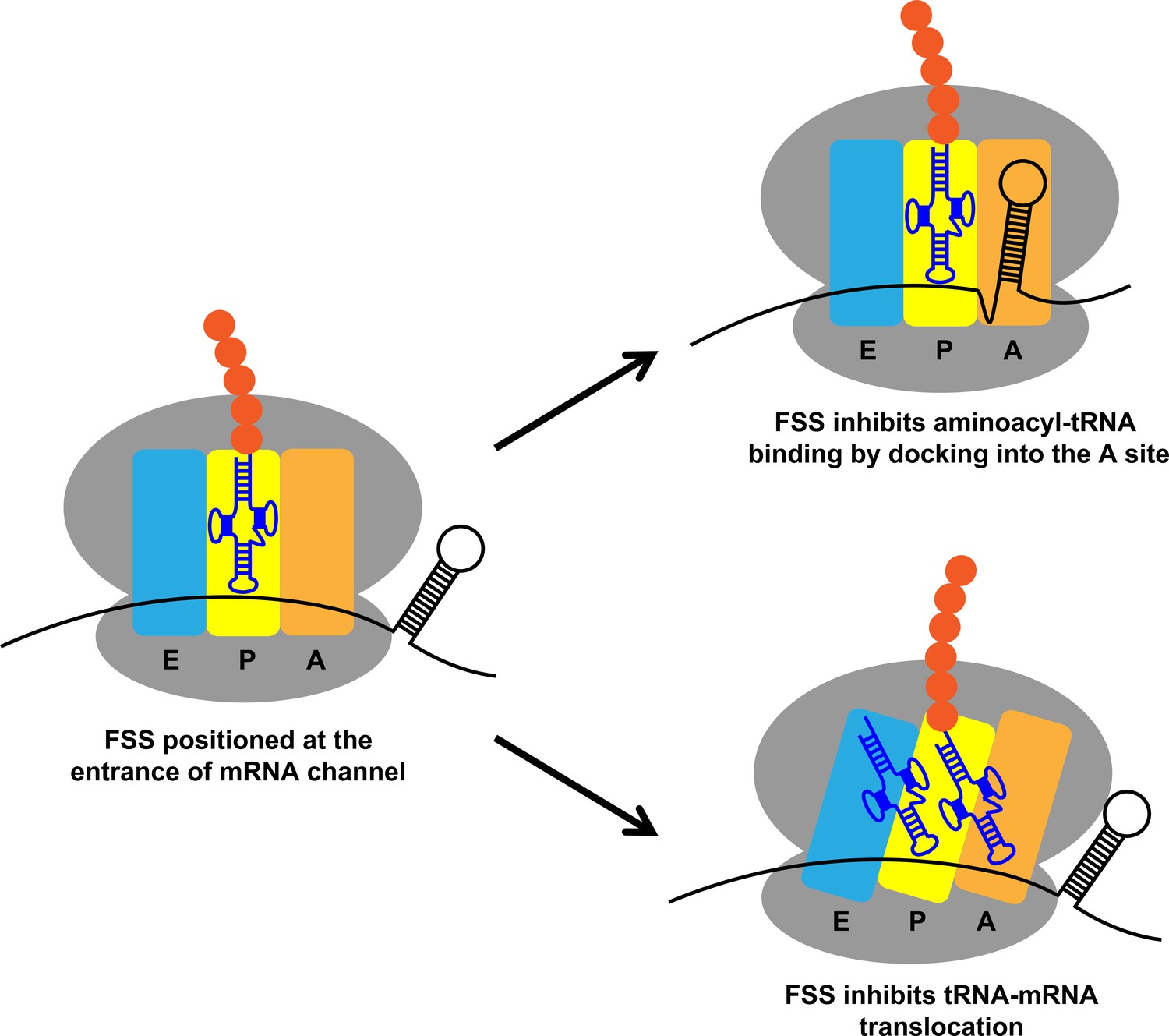
Frontiers | Occlusion of the Ribosome Binding Site Connects the Translational Initiation Frequency, mRNA Stability and Premature Transcription Termination